-Search query
-Search result
Showing all 47 items for (author: tan & cw)

EMDB-16110: 
Human Urea Transporter UT-A (N-Terminal Domain Model)
Method: single particle / : Chi G, Pike ACW, Maclean EM, Mukhopadhyay SMM, Bohstedt T, Scacioc A, Wang D, McKinley G, Fernandez-Cid A, Arrowsmith CH, Bountra C, Edwards A, Burgess-Brown NA, van Putte W, Duerr K

EMDB-16111: 
Map of Human Urea Transporter UT-A Collected with 0 and 30 Degree Tilts
Method: single particle / : Chi G, Pike ACW, Maclean EM, Bohstedt T, Wang D, Mckinley G, Fernandez-Cid A, Mukhopadhyay SMM, Burgess-Brown NA, Edwards A, Arrowsmith C, Bountra C, Duerr KL

EMDB-16112: 
Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14
Method: single particle / : Chi G, Dietz L, Pike ACW, Maclean EM, Mukhopadhyay SMM, Bohstedt T, Wang D, Scacioc A, McKinley G, Arrowsmith CH, Edwards A, Bountra C, Fernandez-Cid A, Burgess-Brown NA, Duerr KL

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-28189: 
SARS-CoV-2 Spike in complex with biparatopic nanobody BP10
Method: single particle / : Pymm PG, Glukhova A, Tham WH
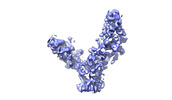
EMDB-28190: 
SARS-CoV-2 RBD in complex with biparatopic nanobody BP10 local refinement
Method: single particle / : Pymm PG, Glukhova A, Tham WH

EMDB-26383: 
Cryo-EM structure of the core human NADPH oxidase NOX2
Method: single particle / : Noreng S, Ota N, Sun Y, Masureel M, Payandeh J, Yi T, Koerber JT

EMDB-15526: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae
Method: electron tomography / : Bieber A, Capitanio C, Schulman BA, Baumeister W, Wilfling F

EMDB-15545: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #2
Method: electron tomography / : Capitanio C, Bieber A, Erdmann PS, Schulman BA, Baumeister W, Wilfling F

EMDB-15546: 
In situ cryo-electron tomogram of a bulk autophagy autophagosome with END cargo in S. cerevisiae #1
Method: electron tomography / : Bieber A, Capitanio C, Erdmann PS, Schulman BA, Baumeister W, Wilfling F

EMDB-15547: 
In situ cryo-electron tomogram of a bulk autophagy autophagosome fusing with the vacuole in S. cerevisiae #1
Method: electron tomography / : Bieber A, Capitanio C, Erdmann PS, Schulman BA, Baumeister W, Wilfling F

EMDB-15548: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #3
Method: electron tomography / : Bieber A, Capitanio C, Erdmann PS, Schulman BA, Baumeister W, Wilfling F

EMDB-15549: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #4
Method: electron tomography / : Capitanio C, Bieber A, Erdmann PS, Schulman BA, Baumeister W, Wilfling F

EMDB-25817: 
Cryo-EM map of protomer of the cytoplasmic ring of the nuclear pore complex from Xenopus laevis
Method: single particle / : Fontana P, Wu H

EMDB-11804: 
Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-11805: 
Structure of Wild-type Human Potassium Chloride Transporter KCC3 in NaCl (MSP E3D1)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-22995: 
Spike protein trimer
Method: single particle / : Asarnow D, Faust B, Bohn M, Bulkley D, Manglik A, Craik CS, Cheng Y

EMDB-12311: 
Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-11799: 
Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map)
Method: single particle / : Chi G, Man H, Ebenhoch R, Reggiano G, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Singh NK, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, DiMaio F, Duerr KL, Structural Genomics Consortium (SGC)

EMDB-11800: 
Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Subclass)
Method: single particle / : Chi G, Man H, Ebenhoch R, Reggiano G, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Singh NK, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, DiMaio F, Duerr KL, Structural Genomics Consortium (SGC)

EMDB-22993: 
SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I
Method: single particle / : Asarnow D, Charles C, Cheng Y

EMDB-22994: 
SARS-CoV-2 Spike protein in complex with Fab 2H4
Method: single particle / : Asarnow D, Charles C, Cheng Y

EMDB-22997: 
SARS-CoV-2 spike glycoprotein:Fab 3D11 complex
Method: single particle / : Asarnow D, Charles C, Cheng Y

EMDB-23707: 
SARS-CoV-2 Spike:5A6 Fab complex I focused refinement
Method: single particle / : Asarnow D, Cheng Y

EMDB-23709: 
SARS-CoV-2 Spike:Fab 3D11 complex focused refinement
Method: single particle / : Asarnow D, Cheng Y

EMDB-23717: 
SARS-CoV-2 S-NTD + Fab CM25
Method: single particle / : Johnson NV, Mclellan JS

EMDB-10704: 
Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl
Method: single particle / : Chi G, Man H, Ebenhoch R, Reggiano G, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Singh NK, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, DiMaio F, Duerr KL

EMDB-11128: 
Cryo-electron tomogram of human Uromodulin filaments
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11129: 
Subtomogram average of human Uromodulin filaments
Method: subtomogram averaging / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11130: 
Cryo-electron tomogram of elastase treated human Uromodulin filaments (eUmod)
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11131: 
Subtomogram average of elastase treated human Uromodulin filaments (eUmod)
Method: subtomogram averaging / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11132: 
Cryo-electron tomogram of human Uromodulin filaments incubated with FimH lectin domain
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R
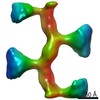
EMDB-11133: 
Subtomogram average of human Uromodulin filaments incubated with FimH lectin domain
Method: subtomogram averaging / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11134: 
Cryo-electron tomogram of elastase treated human Uromodulin filaments incubated with FimH lectin domain
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11135: 
Subtomogram average of elastase treated human Uromodulin fibers incubated with FimH lectin domain
Method: subtomogram averaging / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11136: 
Cryo-electron tomogram of E. coli AAEC [pSH2] incubated with human Uromodulin filaments
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11137: 
Cryo-electron tomogram of non-piliated E. coli AAEC189 incubated with human Uromodulin filaments
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11138: 
Cryo-electron tomogram of FIB milled E.coli - Uromodulin aggregates
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11139: 
Cryo-electron tomogram of E. coli AAEC189 [pSH2] incubated with elastase treated human Uromodulin filaments
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11140: 
Cryo-electron tomogram of urine from patient with E. coli urinary tract infection
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11141: 
Cryo-electron tomogram of urine from patient with K. pneumoniae urinary tract infection
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11142: 
Cryo-electron tomogram of urine from patient with P. aeruginosa urinary tract infection
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-11143: 
Cryo-electron tomogram of urine from patient with S. mitis urinary tract infection
Method: electron tomography / : Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, Trueck J, Devuyst O, Aebi M, Pilhofer M, Glockshuber R

EMDB-4746: 
CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form)
Method: single particle / : Pike ACW, Bushell SR, Shintre CA, Tessitore A, Baronina A, Chu A, Mukhopadhyay S, Shrestha L, Chalk R, Burgess-Brown NA, Love J, Huiskonen JT, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-4747: 
CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form)
Method: single particle / : Pike ACW, Bushell SR, Shintre CA, Tessitore A, Chu A, Mukhopadhyay S, Shrestha L, Chalk R, Burgess-Brown NA, Love J, Huiskonen JT, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)

EMDB-4748: 
CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form)
Method: single particle / : Pike ACW, Bushell SR, Shintre CA, Tessitore A, Chu A, Mukhopadhyay S, Shrestha L, Chalk R, Burgess-Brown NA, Love J, Huiskonen JT, Edwards AM, Arrowsmith CH, Bountra C, Carpenter EP, Structural Genomics Consortium (SGC)
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
